Marie Delattre, a biologist at the École Normale Supérieure de Lyon, has been studying the nematode *Mesorhabditis belari* for nearly a decade. Her initial interest in this microscopic worm stemmed from its unique reproductive strategy, in which only a small fraction of offspring retain their male parent's DNA, while the rest rely solely on maternal genes. However, Delattre's focus shifted when she made an unexpected observation under the microscope.
While examining *Mesorhabditis* embryos, she noticed an unusual phenomenon: in some of the embryos, the DNA appeared shattered into small fragments. This piqued her curiosity, prompting a closer look across different stages of embryo development. At the one-cell stage, the DNA remained intact, looking entirely normal. It continued to remain stable as the embryo transitioned from a single cell to two cells, and then to three. But at the five-cell stage, a striking change occurred—the DNA fragments suddenly emerged.
These DNA fragments first appeared within the nucleus, then dispersed into the surrounding cytoplasm. As cell division continued, they eventually vanished entirely from the embryos. Delattre was fascinated by this transient phase in the worm’s development, during which part of the genome seemed to disappear.
This phenomenon is known as *programmed DNA elimination*, a process seen in dozens of other species where specific parts of the genome are intentionally removed during development. "It was a really random discovery," Delattre remarked. "Serendipity, I would say." Her team's subsequent investigation, recently published in the journal *Current Biology*, expanded the understanding of this enigmatic process, adding *Mesorhabditis* nematodes to the growing list of species capable of programmed DNA elimination. This discovery opens new avenues for understanding how organisms can selectively manage their genetic material during development.
Despite over a century of research, programmed DNA elimination remains a mystery with many unanswered questions. Jianbin Wang, a biologist at the University of Tennessee, Knoxville, highlights the significance of this phenomenon: *"You see that this thing happens very commonly, suggesting it has an important biological role. It's just that we don't really know what that role is yet."* This phenomenon, where certain segments of DNA are systematically removed from cells, continues to intrigue scientists worldwide.
Many researchers, including those like Delattre, have stumbled upon this process unexpectedly. Yet, their initial curiosity has evolved into a dedicated pursuit, as they recognize that understanding programmed DNA elimination could reshape our knowledge of genome dynamics. Their efforts are being bolstered by advancements in molecular biology and genomic technologies, which allow for a deeper exploration of these processes than ever before.
The roots of this research trace back to the 1880s at the Zoological Institute, where cell biologist Theodor Boveri studied *Parascaris*, a species of parasitic worm with a remarkably large genome. Using the limited tools of his time, Boveri observed that significant portions of the germline genome were discarded during the development of somatic cells. This early observation hinted at the possibility of DNA elimination playing a role in cellular differentiation.
Fast forward over 100 years, and modern molecular techniques have confirmed Boveri’s findings in greater detail. Studies have revealed that *Parascaris* worms eliminate a staggering 89 percent of their 2.5-billion-base germline genome as their somatic cells develop. This drastic DNA reduction highlights the potential importance of programmed DNA elimination in regulating the development and function of organisms. The ongoing research could unlock a deeper understanding of genome plasticity and its role in evolution, development, and possibly disease mechanisms.
In the early days of cell biology, Boveri observed programmed DNA elimination, initially believing it to be a normal aspect of development. However, as researchers investigated this process in a broader range of organisms, they realized that it was not a universal phenomenon. Early research concentrated on microscopic species, including parasitic worms and single-celled organisms known as ciliates. A significant breakthrough came from studying the parasitic worm family *Ascaris*, which eliminates approximately one-fifth of its germline genome during development. This work provided crucial insights into the nature of programmed DNA elimination.
It wasn't until the 1980s that scientists identified a family of vertebrates that also undergoes this process: hagfish, which eliminate between 20% and 50% of their germline genome. More recent studies have revealed that nearly all songbirds appear to eliminate parts of their germline genomes as well.
"We are exposed to organisms that have programmed DNA elimination every single day," noted Alexander Suh, an evolutionary biologist at the Leibniz Institute for the Analysis of Biodiversity Change and Uppsala University. Advances in DNA sequencing technology have greatly enhanced researchers' ability to study this phenomenon. By comparing the genomic sequences of germ cells and somatic cells within the same organism, scientists can identify long segments present in the germline genome but absent from the somatic genome. These studies have demonstrated that various species can eliminate anywhere from 0.5% to 90% of their genomes, revealing the diverse scope and evolutionary significance of this process.
Scientists have utilized genome sequencing to precisely determine which regions of DNA are removed during cellular processes and what genetic instructions these regions contain. Generally, the same DNA regions are excised in every cell of a particular organism and are consistent across individuals within a species, though variations exist between different species. Despite these differences, the regions that are eliminated, irrespective of the species, tend to consist of large segments of repeated DNA sequences. These repetitive sequences typically do not carry instructions for protein synthesis, suggesting that their removal may be part of a regulatory or non-coding function in the genome. Understanding this selective DNA removal can provide insights into genetic regulation, evolution, and the functional significance of non-coding DNA across different organisms.
Understanding Programmed DNA Elimination
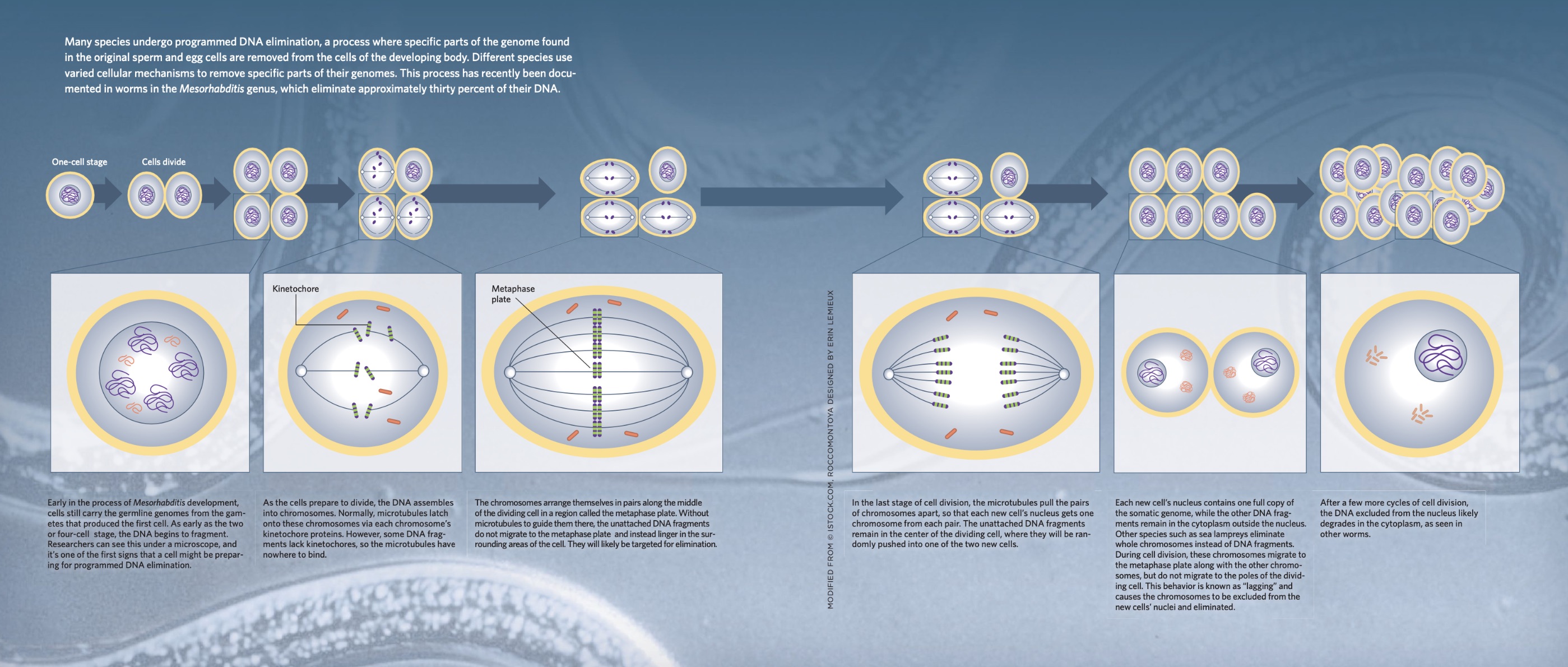
Programmed DNA elimination is a phenomenon that occurs across various branches of the tree of life, with diverse methods observed among different organisms. From nematodes to unicellular ciliates, the process can involve slicing genomes into small fragments and removing specific segments. In contrast, vertebrates tend to eliminate entire chromosomes. Despite these variations, the core steps of DNA elimination are similar: certain portions of the genome are marked for removal, and during cell division, this DNA is segregated out of the nucleus and eventually expelled from the cell.
In organisms like ciliates and nematodes, the process involves fragmenting the DNA before removal, but how this happens at the cellular level remains largely unclear. However, research by scientists such as Wang and Delattre is beginning to shed light on the mechanics of this process.
The Role of Cell Division in DNA Elimination
Cell division, a precise and well-regulated process, plays a crucial role in DNA elimination. During division, duplicated chromosomes align along the metaphase plate at the cell's center. Microtubules, extending from opposite poles of the cell, attach to kinetochore proteins located at the center of each chromosome. These microtubules then pull half of the chromosomes into each new cell.
In the nematode *Ascaris*, research by Wang has shown that DNA fragments destined for elimination lack the necessary kinetochore proteins. Without these proteins, the microtubules cannot attach to the fragments, preventing their incorporation into the newly formed nuclei. Similarly, Delattre observed this absence of kinetochore proteins in the DNA fragments of *Mesorhabditis*. Using fluorescent markers, she visualized these DNA fragments as tiny dots encircling the remaining chromosomes. As cell division progresses, these untethered DNA fragments are pushed outward into the cytoplasm, where they eventually disappear.
DNA Elimination in Vertebrates
In vertebrate species, where entire chromosomes are often eliminated, the process diverges slightly. Problems arise during cell division when certain chromosomes, known as "lagging" chromosomes, move more slowly than others. This lagging is particularly evident in sea lampreys, where a piece of DNA at the tip of the chromosome appears to pull the chromosome away from the direction of the microtubules. As a result, these lagging chromosomes are left behind when the new nuclei form, eventually degrading in the cytoplasm.
Unanswered Questions and Ongoing Research
The mechanisms that determine which DNA fragments or chromosomes are selected for elimination remain a mystery. In her research, Delattre is working to map the genome in detail and measure RNA expression to understand how cells identify which DNA to fragment and which proteins are responsible for the cuts. She aims to identify specific protein candidates involved in the process, with plans to experimentally remove these proteins from embryos to observe if programmed DNA elimination still occurs.
As researchers continue to unravel these complex processes, the study of programmed DNA elimination offers insights into fundamental aspects of genome regulation and evolution, revealing the intricate dance between cellular machinery and genetic material.
Extreme DNA silencing
Here's a rephrased and expanded version of the content, incorporating additional relevant information:
A key question in the study of programmed DNA elimination is: why would a species choose to eliminate parts of its DNA at all? According to Jeramiah Smith, a biologist at the University of Kentucky, removing DNA is an extreme strategy for preventing certain genes from being used in cells. Cells have less disruptive methods for achieving the same outcome, such as tightly packing their DNA so that specific genes become inaccessible, or using small RNA molecules that bind to genes and inhibit protein production. Interestingly, species that undergo programmed DNA elimination usually possess these mechanisms to silence genes as well.
Another hypothesis is that the eliminated genes might play specific roles in reproductive cells (germ cells), where they may be necessary to produce proteins that are not required in other cells of the body. Research on species like *Ascaris* worms and zebra finches has shown that the genes eliminated during this process are often active in reproductive organs, such as the testes, where germ cells are formed.
However, not all researchers agree with this perspective. For instance, Delattre’s studies on the nematode *Mesorhabditis* suggest that the genes being eliminated appear to follow no consistent pattern and don’t seem to be essential for the species' survival. She proposes that, in these worms, the process may mainly serve to remove repetitive DNA sequences, with any eliminated genes being incidental.
There is also the possibility that programmed DNA elimination serves different purposes across various species. "At this point, I would say these are distinct processes," noted Suh. Smith concurred, emphasizing that the evolutionary origins of DNA elimination remain unclear. He speculates that different branches of life may have independently developed this ability, adapting it through diverse evolutionary paths. "They're achieving similar outcomes, but through very different evolutionary routes," he remarked.
While programmed DNA elimination hasn't been directly observed in humans, analogous processes can occur when human cells malfunction. "In a normal, healthy cell, you would never see this," Delattre explained. "Such an event would be a major red flag." For example, when human DNA breaks apart in a process called chromothripsis, it leads to a catastrophic cellular event that can result in cancer. Similarly, improper division of chromosomes during early embryonic development can lead to developmental disorders.
Understanding the origins and mechanisms of DNA elimination could shed light on the functioning of our own genome. Smith suggests that this knowledge could improve our understanding of how similar events occur in cancer cells. Moreover, insights into how DNA is naturally marked for elimination could potentially inform new therapeutic approaches, such as selectively deleting excess chromosomes in conditions like Down syndrome.
Looking ahead, many scientists agree that the next step is to investigate programmed DNA elimination across a broader range of species. "We likely don’t yet know the full scope of species that utilize this process," Smith said. Suh and his team are already advancing this research, having sequenced germline and somatic DNA from around 30 species of songbirds. They discovered that every songbird species eliminates at least one chromosome, with the size of the eliminated chromosome varying—sometimes the largest, sometimes the smallest. Interestingly, while the eliminated genes differ greatly between species, they often share similarities with sequences found on retained chromosomes, a trend not yet observed in other animal groups. This finding could provide new insights into the evolutionary significance of DNA elimination.






















0 Comments